Note
Go to the end to download the full example code
Manipulation of UV-Vis data¶
After you’ve looked at the simple UV-Vis example, this one shows how you can manipulate UV-Vis data.
the experiments present in this file are: dict_keys(['rxn buffer w_o', 'rxn buffer with', 'TCM w_o', 'TCMI36C w_o', 'rxn buff w_ellman', 'elution buff w_ellman', 'TCM w_ellman', 'TCMI36C_w_ellman'])
now I'm going to try a DSW file
the experiments present in this file are: dict_keys(['protein_1', 'protein_2', 'protein_3', 'protein_4', 'protein_5', 'protein_6', 'protein_7', 'protein_8', 'protein_9'])
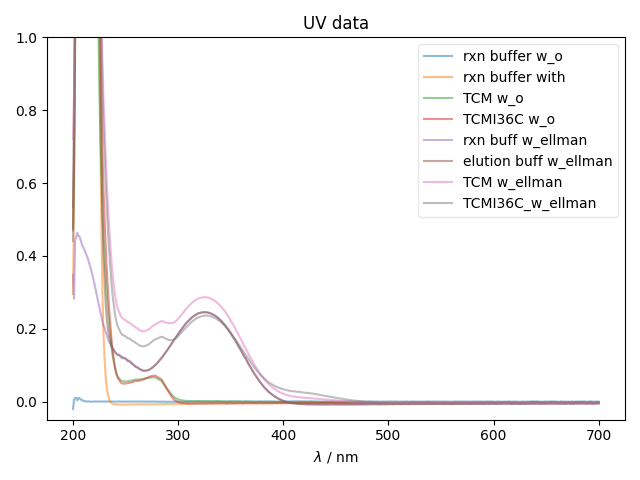
1: UV data |||nm
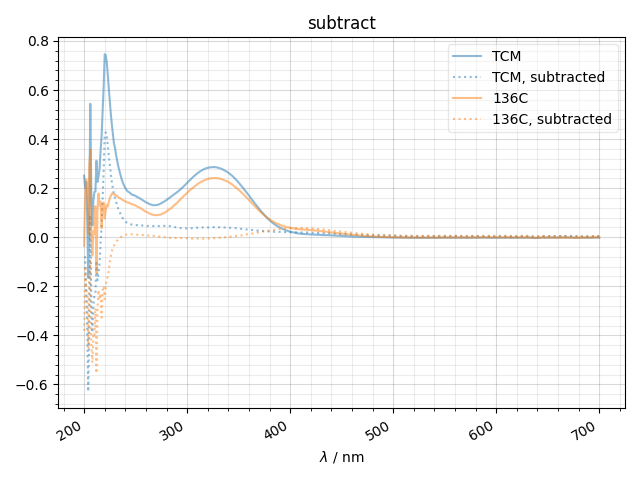
2: subtract |||nm
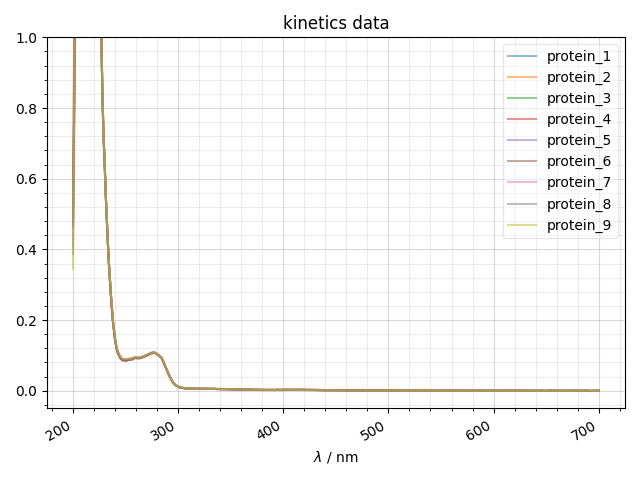
3: kinetics data |||nm
from pylab import *
from pyspecdata import *
from itertools import cycle
color_cycle = cycle(['#1f77b4', '#ff7f0e', '#2ca02c',
'#d62728', '#9467bd', '#8c564b', '#e377c2',
'#7f7f7f', '#bcbd22', '#17becf'])
#init_logging('debug')
data = find_file('200703_Ellman_before_SL.DSW',
exp_type='UV_Vis/Ellmans_Assay')
print("the experiments present in this file are:",data.keys())
with figlist_var() as fl:
fl.next("UV data")
for k,thisspectrum in data.items():
fl.plot(thisspectrum,
alpha=0.5,
label=k)
ylabel(thisspectrum.get_units())
ylim((-0.05,1))
fl.next('subtract')
subdata = {'TCM':data['TCM w_ellman'] - data['TCM w_o'],
'136C':data['TCMI36C_w_ellman'] - data['TCMI36C w_o'],
}
for k,d in subdata.items():
thiscolor = next(color_cycle)
fl.plot(d,
alpha=0.5,
color=thiscolor,
label=k)
fl.plot(d - data['rxn buff w_ellman'],
':',
alpha=0.5,
color=thiscolor,
label='%s, subtracted'%k)
ylabel(d.get_units())
gridandtick(gca())
print("now I'm going to try a DSW file")
data = find_file('Ras_Stability4',
exp_type='UV_Vis/Ras_stability/200803_RT')
print("the experiments present in this file are:",data.keys())
fl.next("kinetics data")
for k,thisspectrum in data.items():
fl.plot(thisspectrum,
alpha=0.5,
label=k)
ylabel(thisspectrum.get_units())
ylim((-0.05,1))
gridandtick(gca())
Total running time of the script: (0 minutes 2.648 seconds)