Note
Go to the end to download the full example code
Calculation of the Covariance Matrix¶
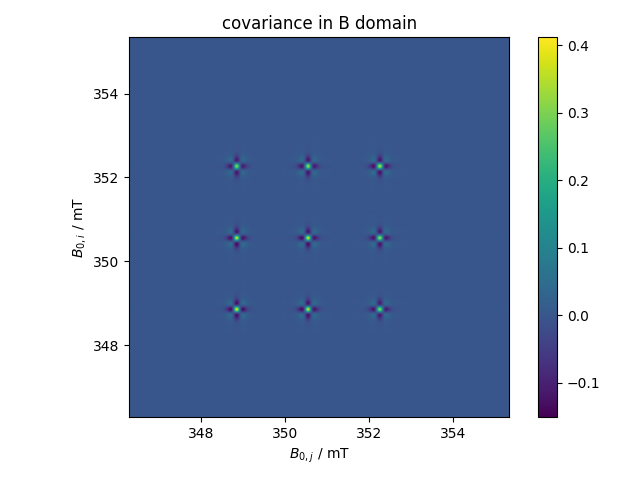
After rescaling plots, the covariance matrix is calculated and then plotted for a 2D Field experiment (spectra as a function of field with multiple collections or “Times”)
1: covariance in B domain |||('mT', 'mT')
2: Covariance in U domain |||('kcyc · (T)$^{-1}$', 'kcyc · (T)$^{-1}$')
from pyspecdata import *
from pylab import *
fieldaxis = "$B_0$"
exp_type = "francklab_esr/alex"
with figlist_var() as fl:
for filenum, (thisfile) in enumerate(
[("230504_3p8mM_TEMPOL_stb_wt_4x.DSC")]
):
d = find_file(thisfile, exp_type=exp_type)["harmonic", 0]
d.set_units(fieldaxis, 'T').setaxis(fieldaxis, lambda x: x*1e-4)
d.rename("Time", "observations")
d.reorder(["observations", fieldaxis])
fl.next("covariance in B domain")
# we do this first, because if we were to ift to go to u domain and
# then ft back, we would introduce a complex component to our data
fl.image(d.C.cov_mat("observations"))
d.ift(fieldaxis, shift=True)
fl.next("Covariance in U domain")
fl.image(d.cov_mat("observations")) # this time, do not spin up an extra copy of the data
Total running time of the script: (0 minutes 25.686 seconds)