Note
Go to the end to download the full example code
Verify the pulse calibration¶
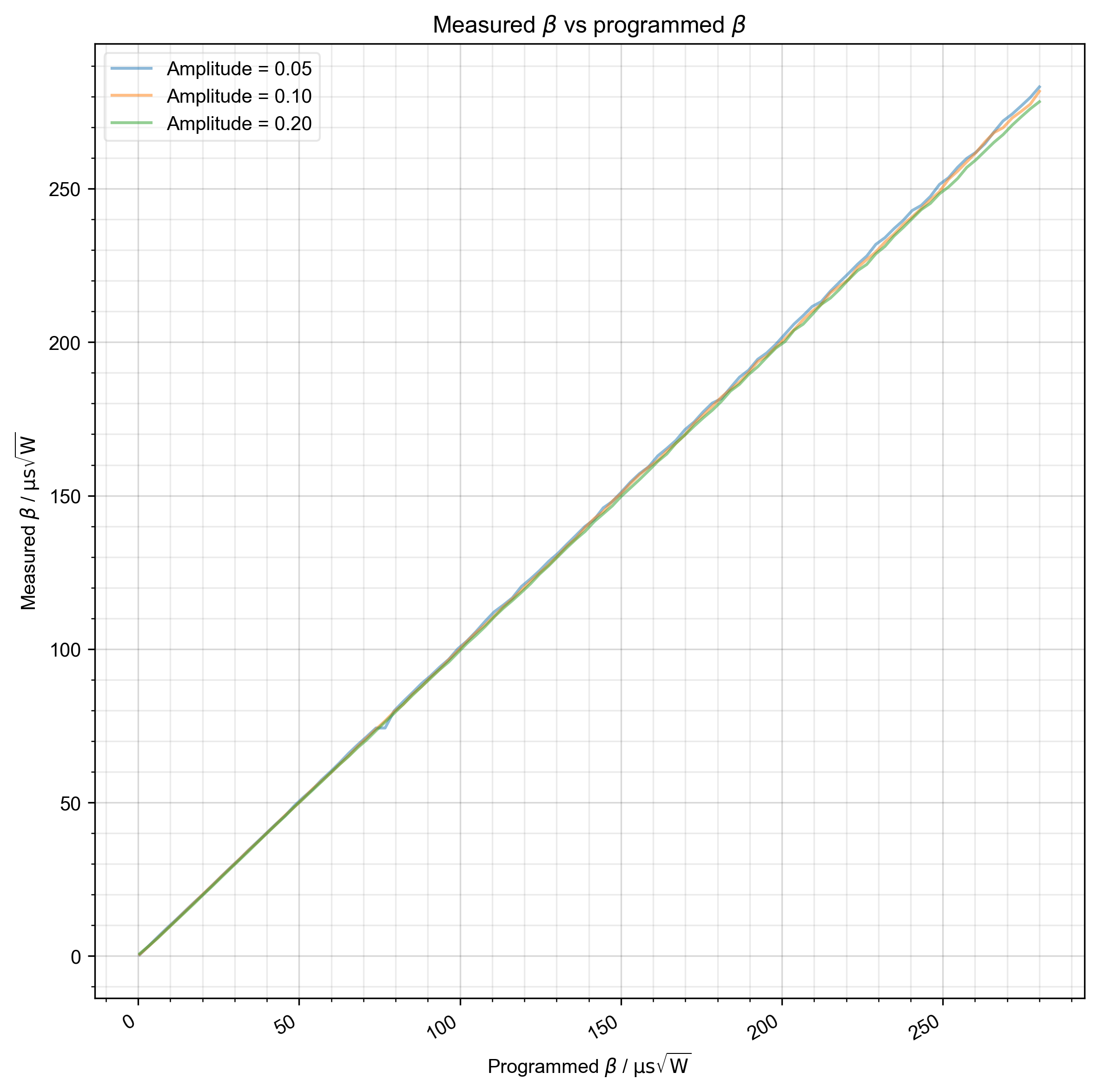
Once we have the pulse-length calibration in place, we want to simply verify that we get the β values that we program, where \(\beta = \frac{1}{\sqrt{2} \int \sqrt{P(t)_{amp}} dt\)
This data should be acquired using example/run_pulse_calibration.py in FLInst.
Compare to example/calibrate_tsqrtP.py, which generates the calibration.
---------- logging output to /home/jmfranck/pyspecdata.0.log ----------
You didn't set units for beta before saving the data!!!
--> simple_functions.py(126):root find_apparent_anal_freq 2024-10-26 15:48:51,629
INFO: Aliasing occurred, but we can still find that frequency!
You didn't set units for beta before saving the data!!!
You didn't set units for beta before saving the data!!!
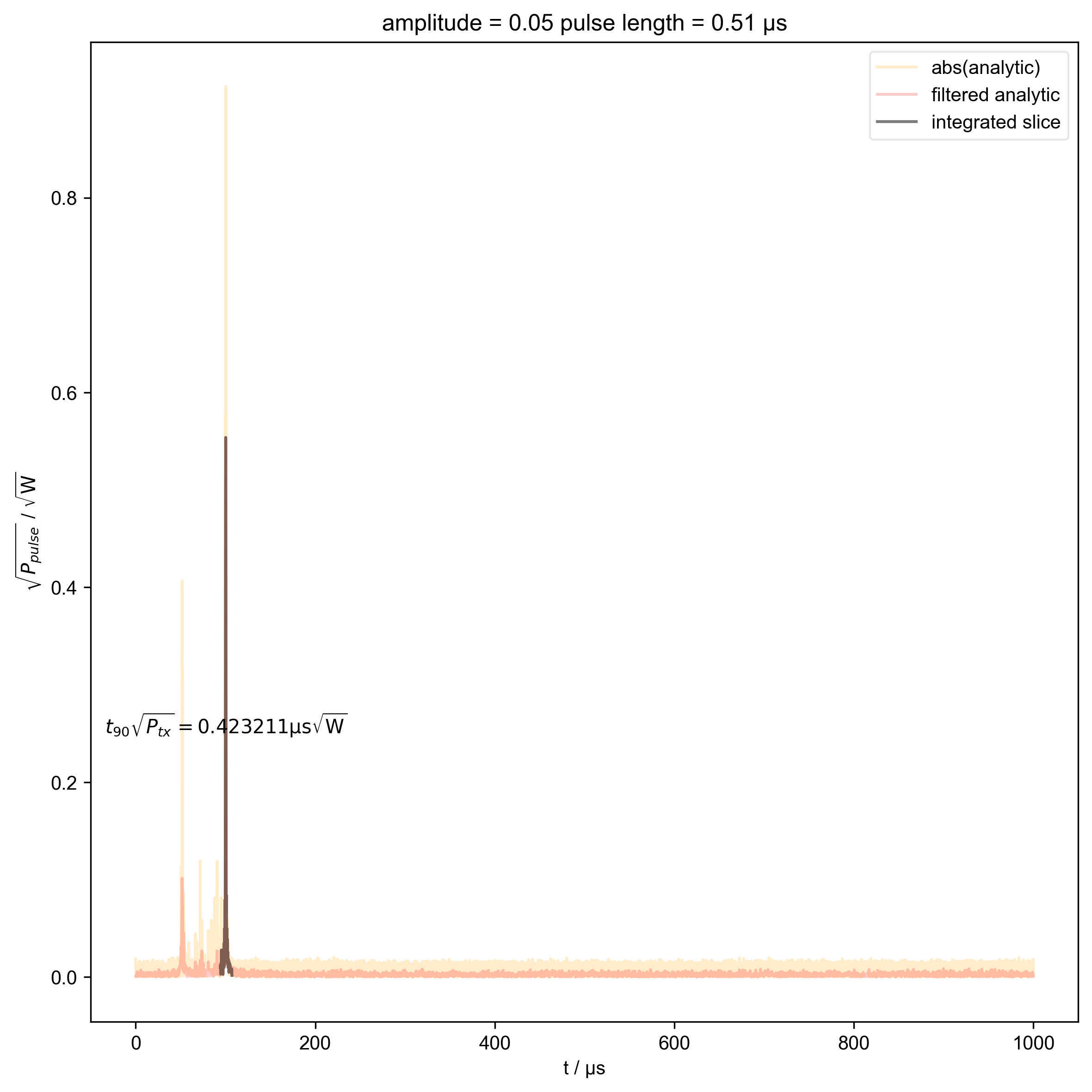
1: amplitude = 0.05 pulse length = 0.51 μs |||μs
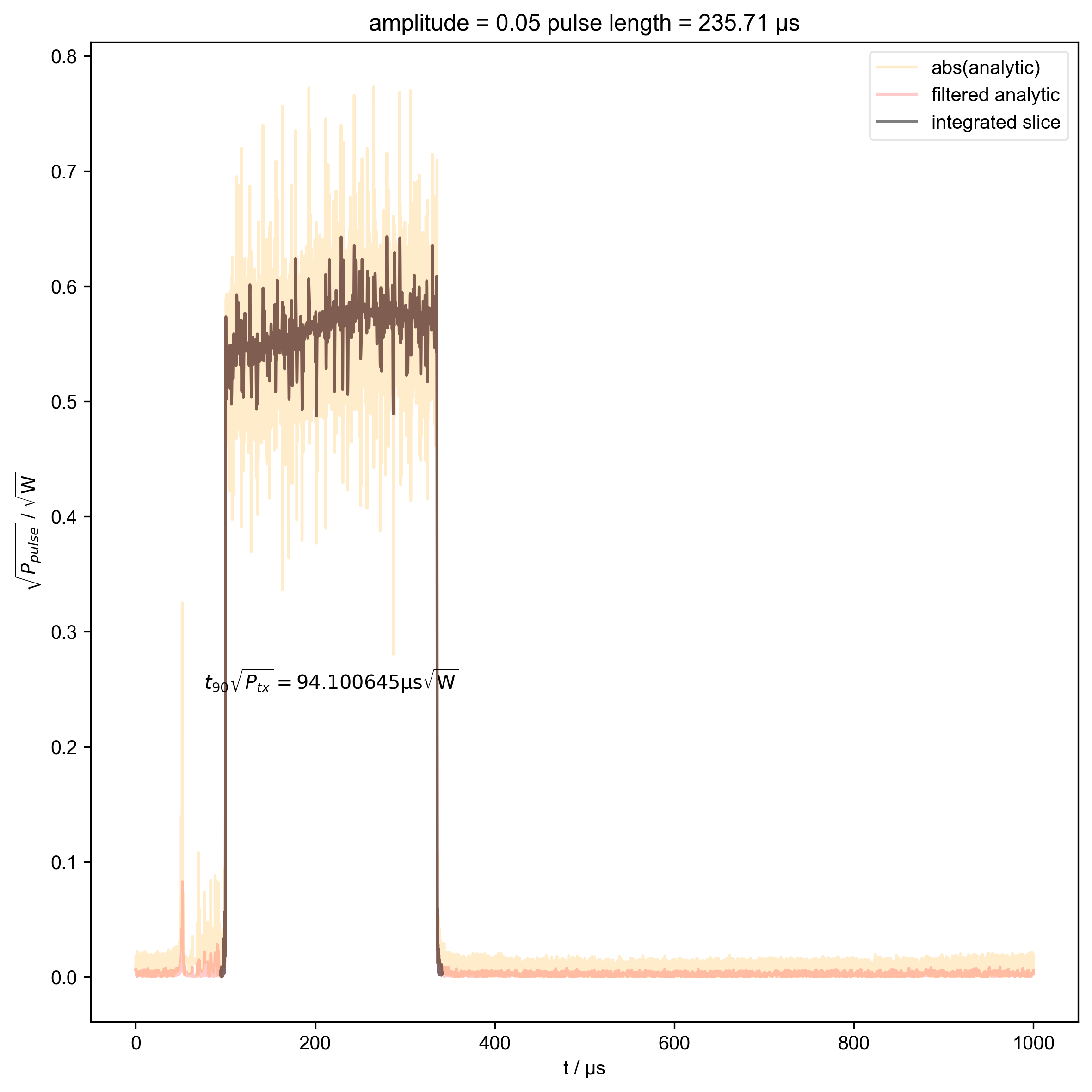
2: amplitude = 0.05 pulse length = 235.71 μs |||μs
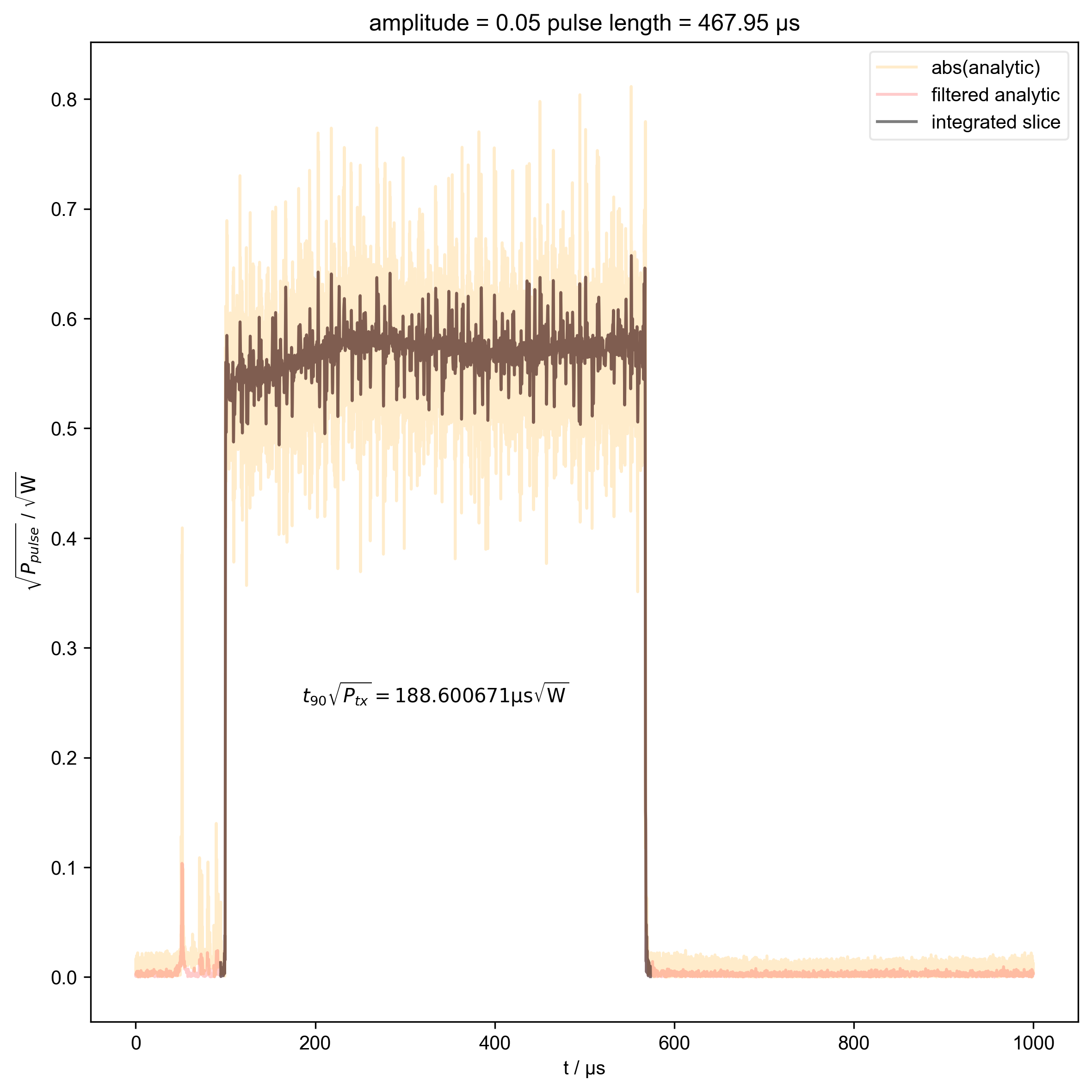
3: amplitude = 0.05 pulse length = 467.95 μs |||μs
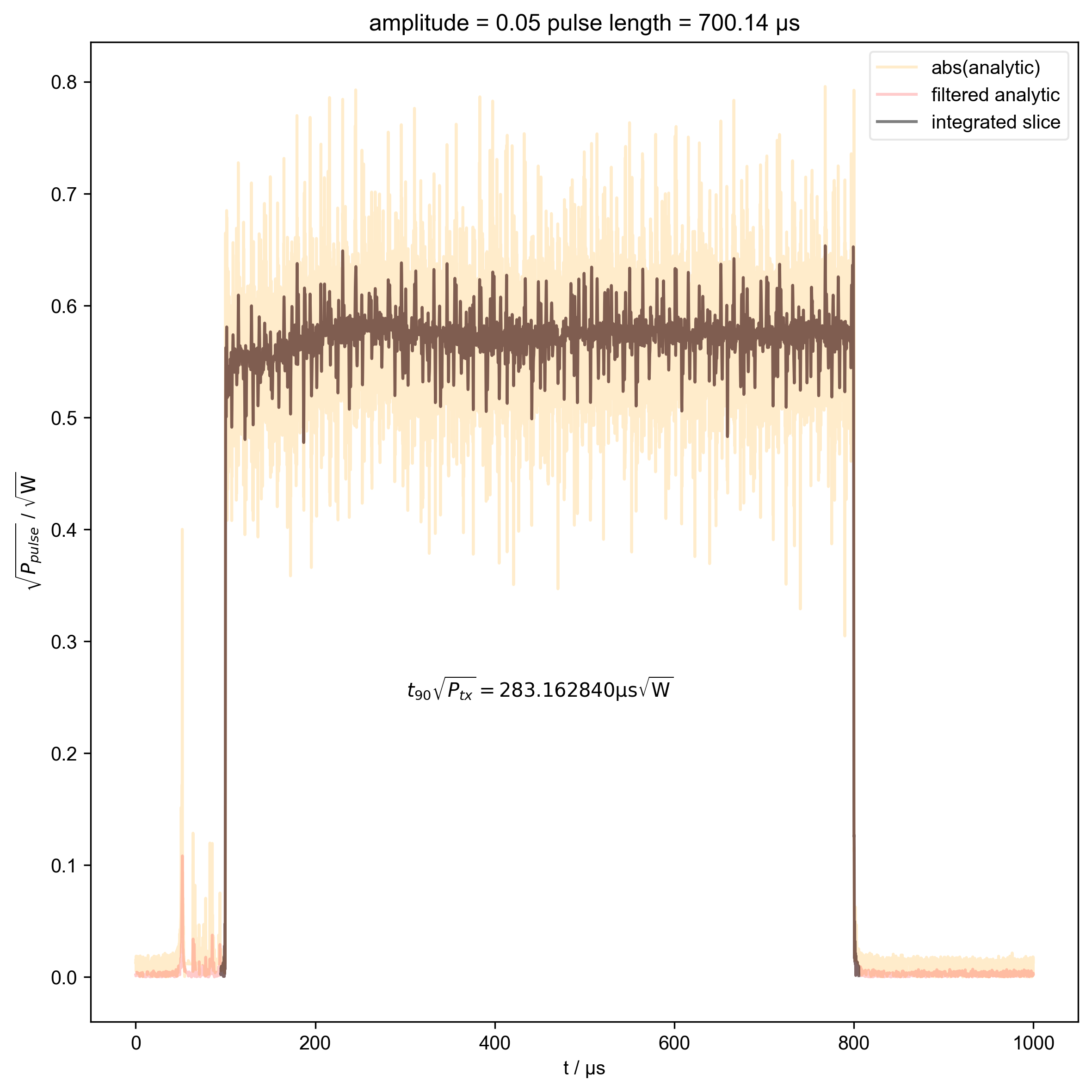
4: amplitude = 0.05 pulse length = 700.14 μs |||μs
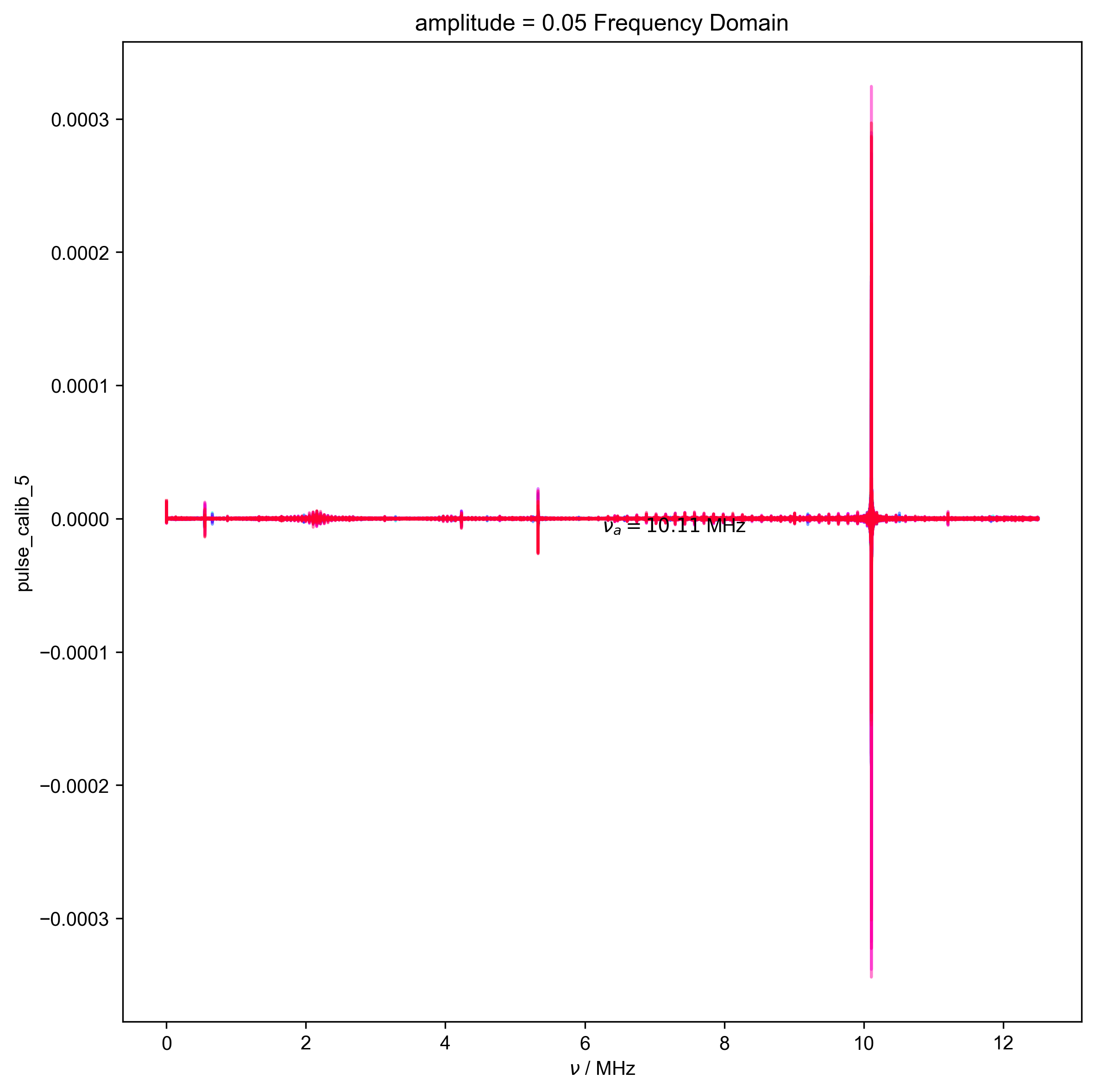
5: amplitude = 0.05 Frequency Domain |||('MHz', None)
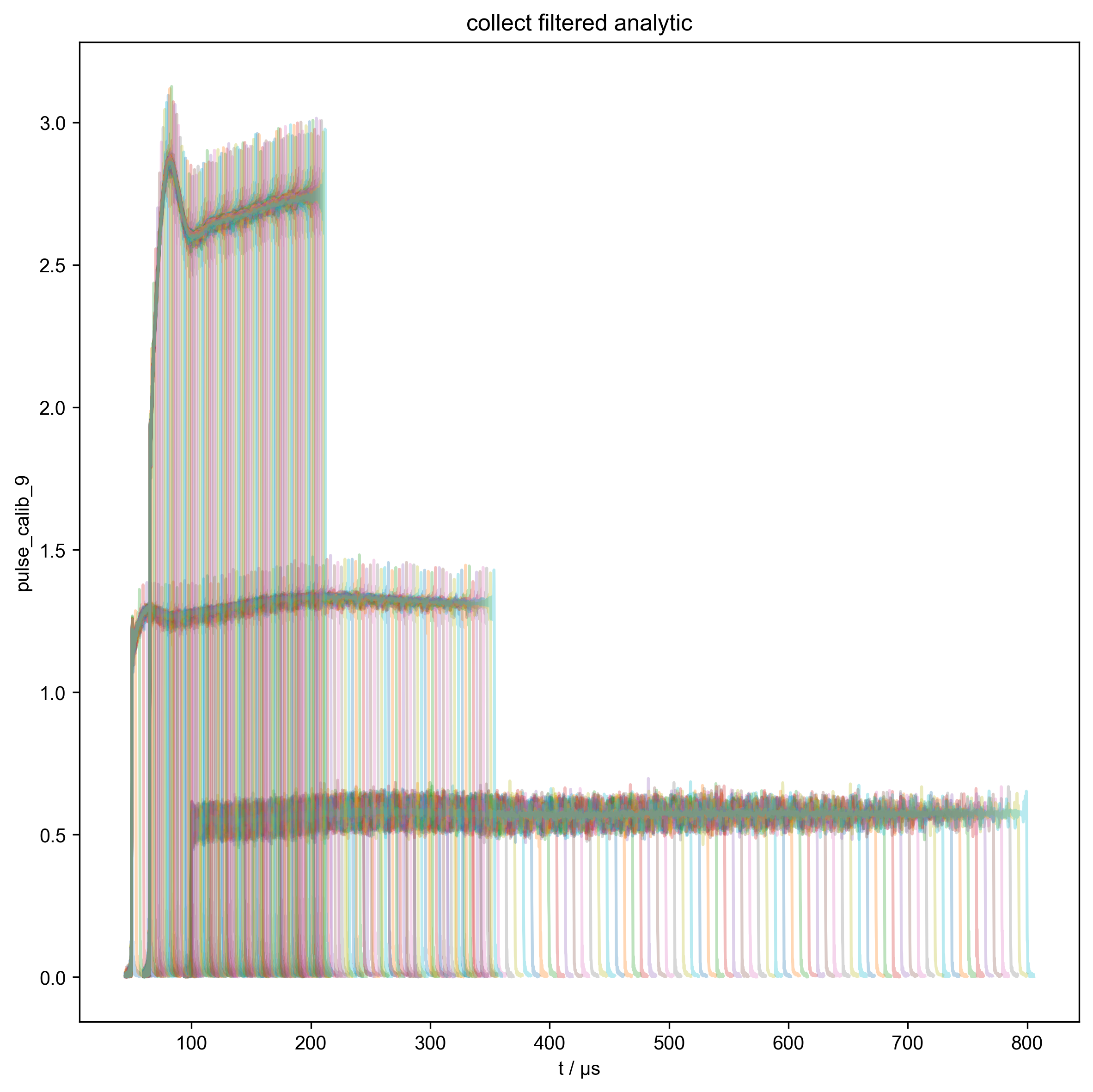
6: collect filtered analytic |||μs
7: Measured $\beta$ vs programmed $\beta$ |||μs√W
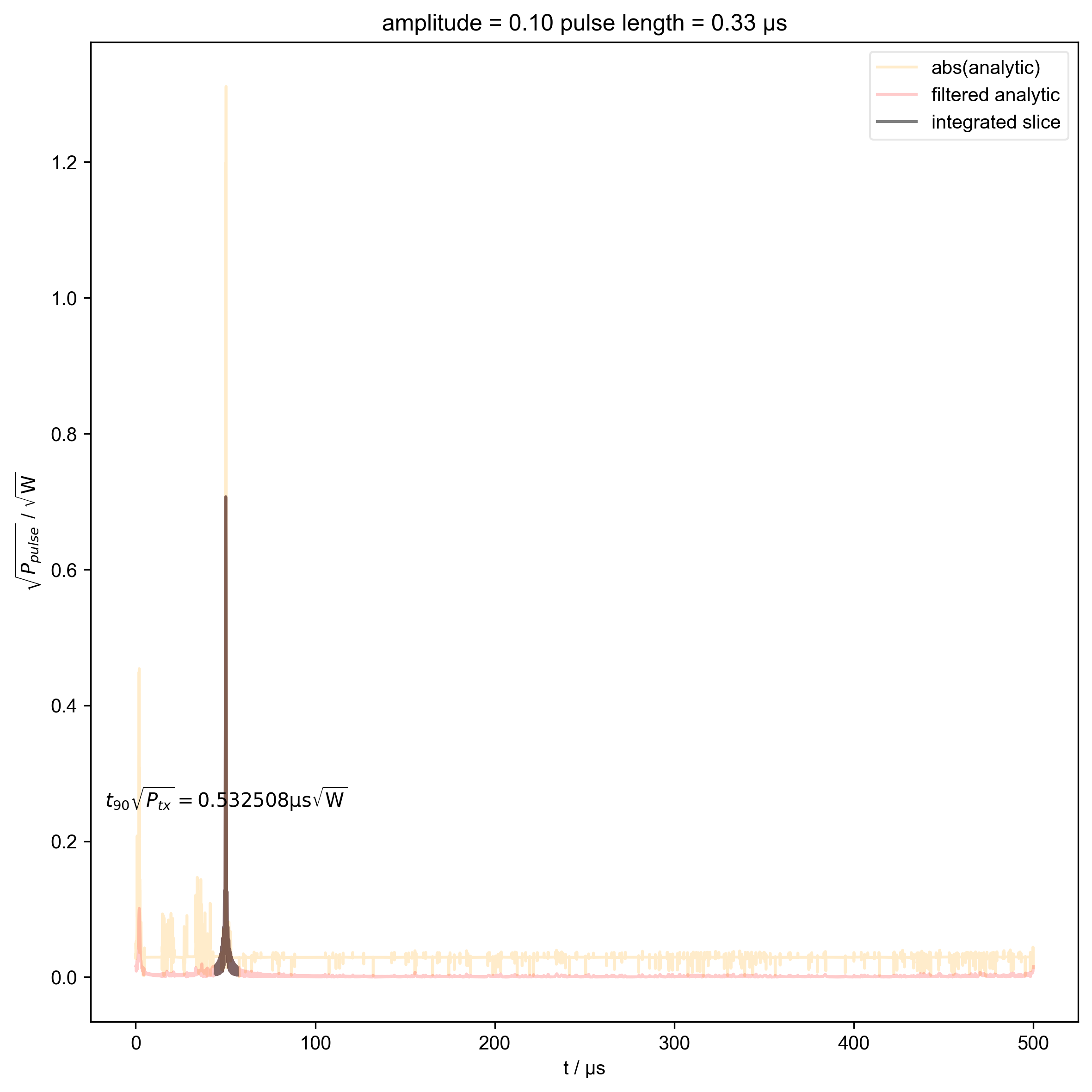
8: amplitude = 0.10 pulse length = 0.33 μs |||μs
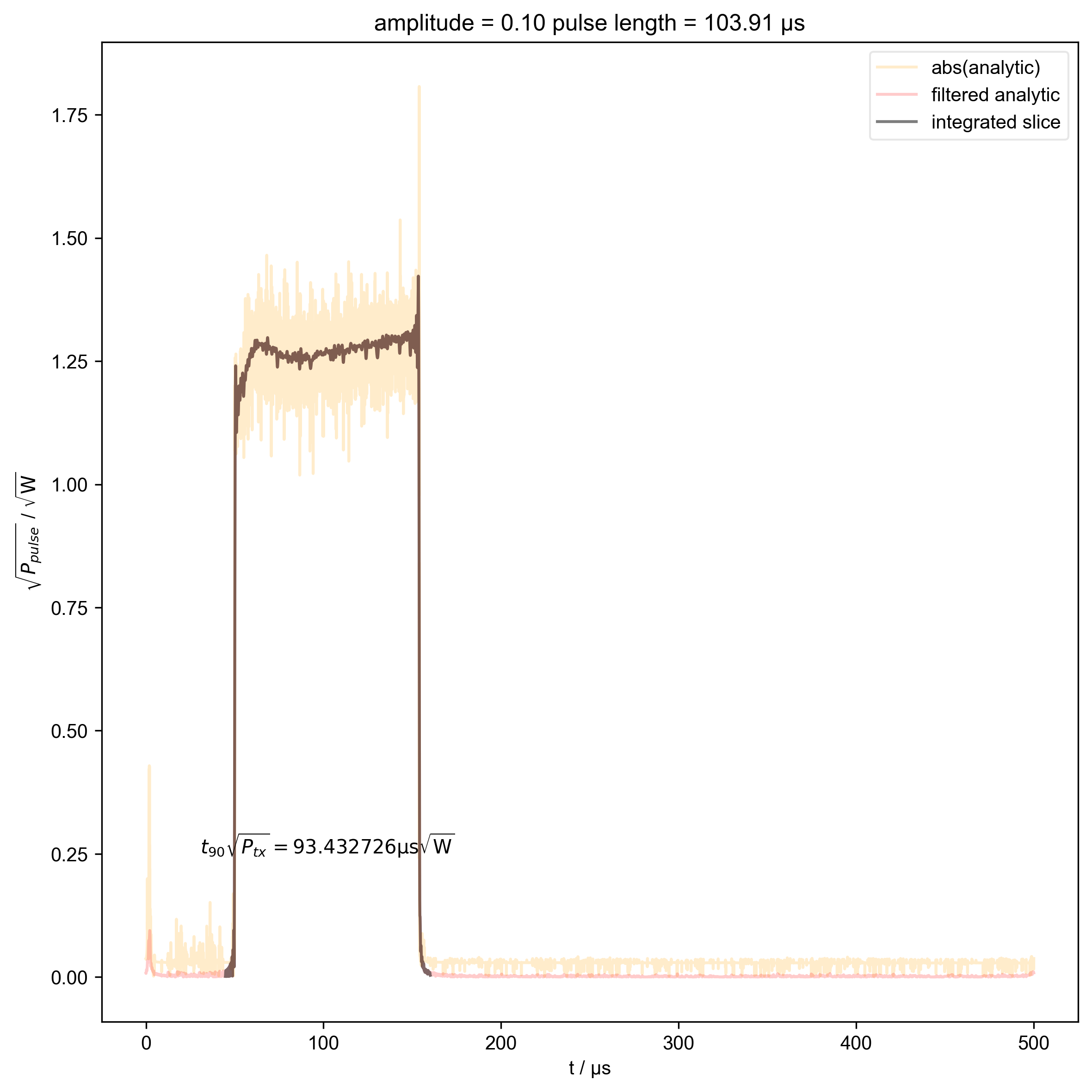
9: amplitude = 0.10 pulse length = 103.91 μs |||μs
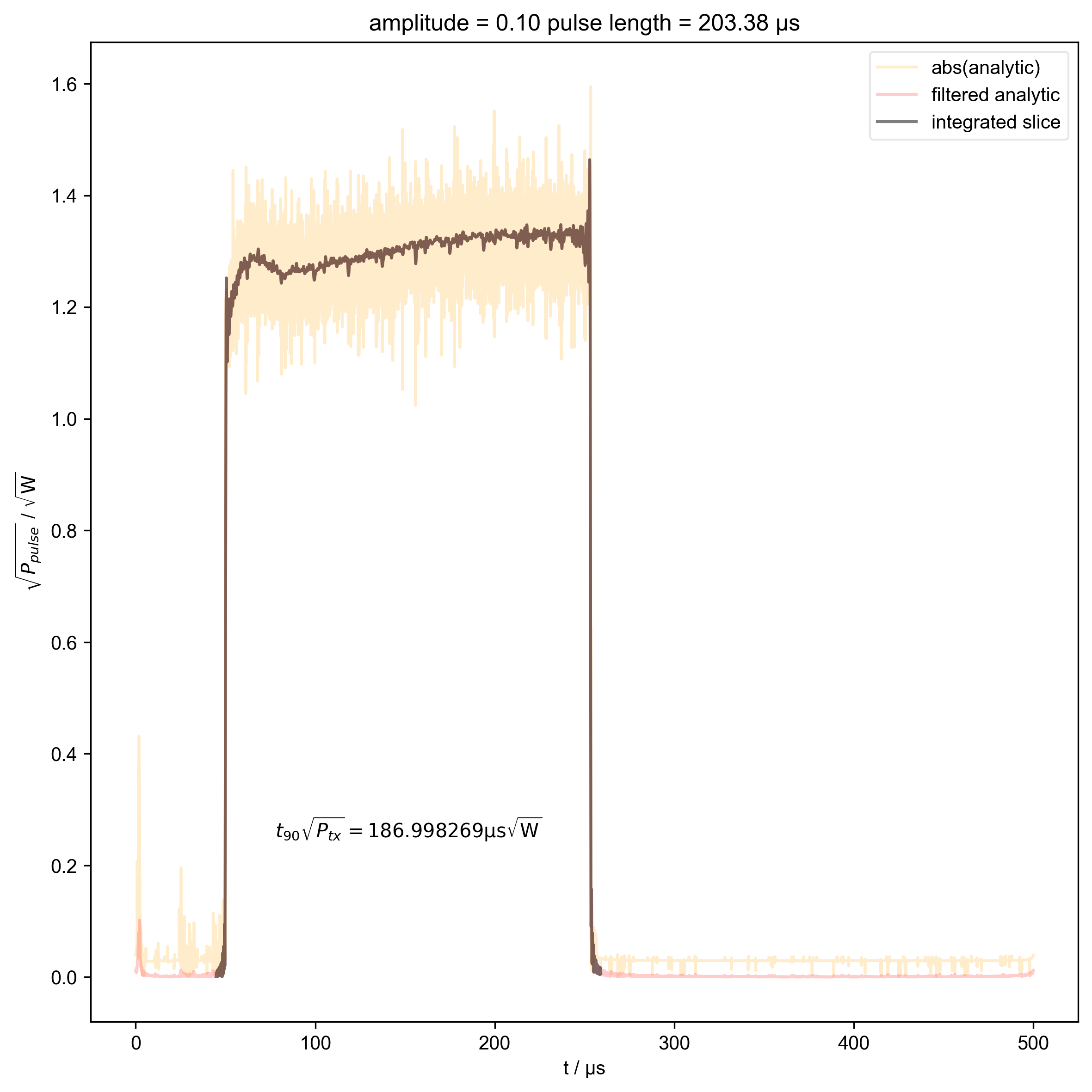
10: amplitude = 0.10 pulse length = 203.38 μs |||μs
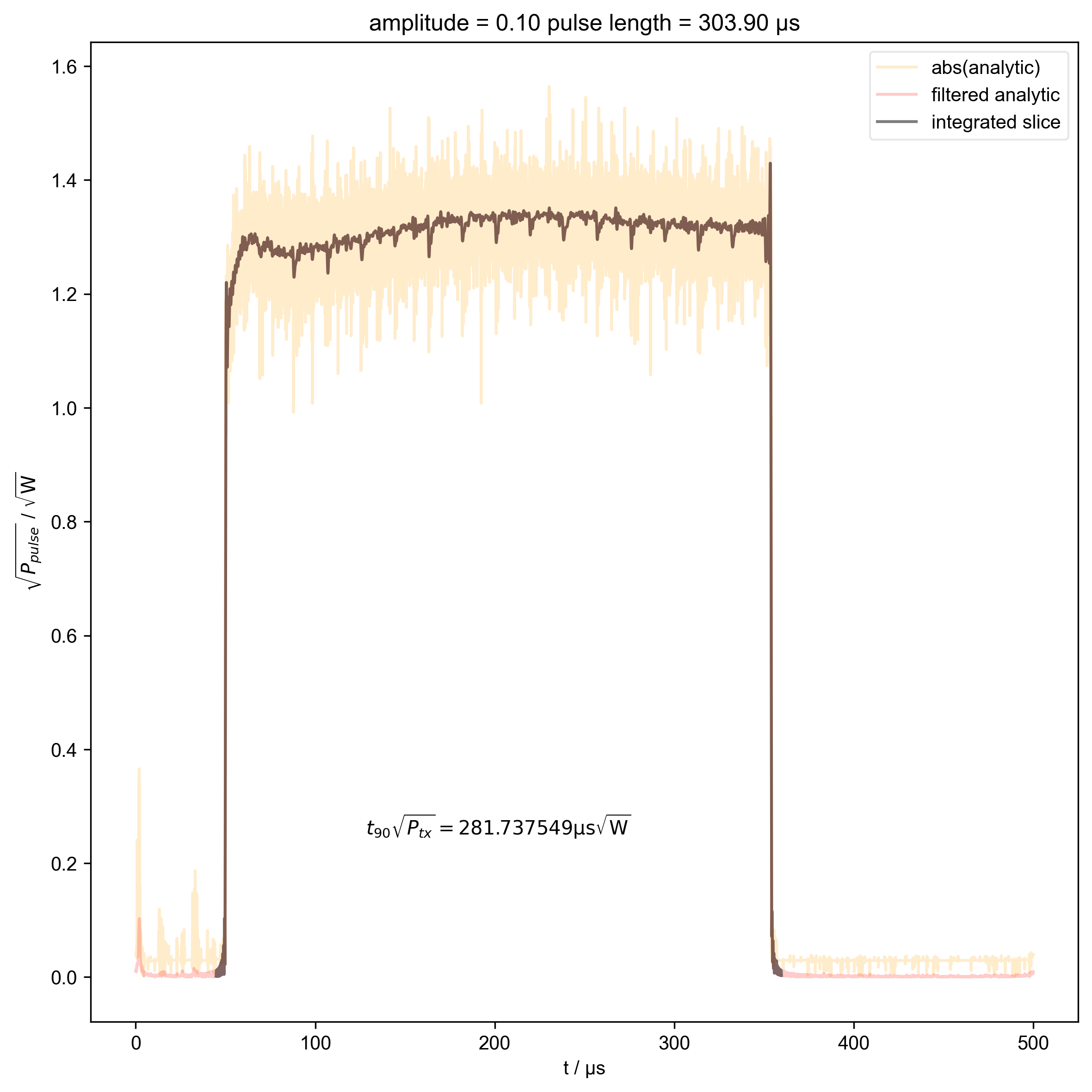
11: amplitude = 0.10 pulse length = 303.90 μs |||μs
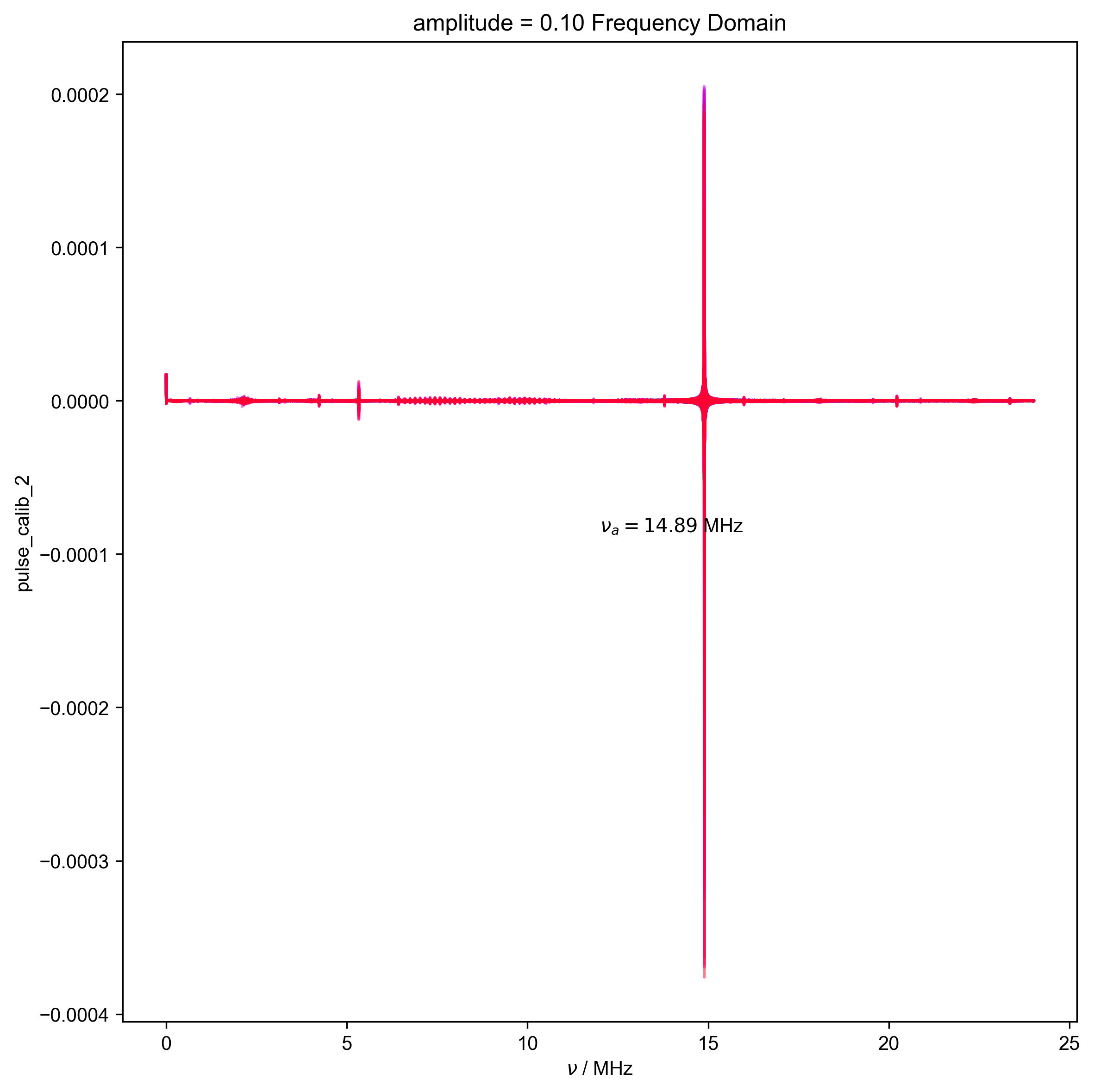
12: amplitude = 0.10 Frequency Domain |||('MHz', None)
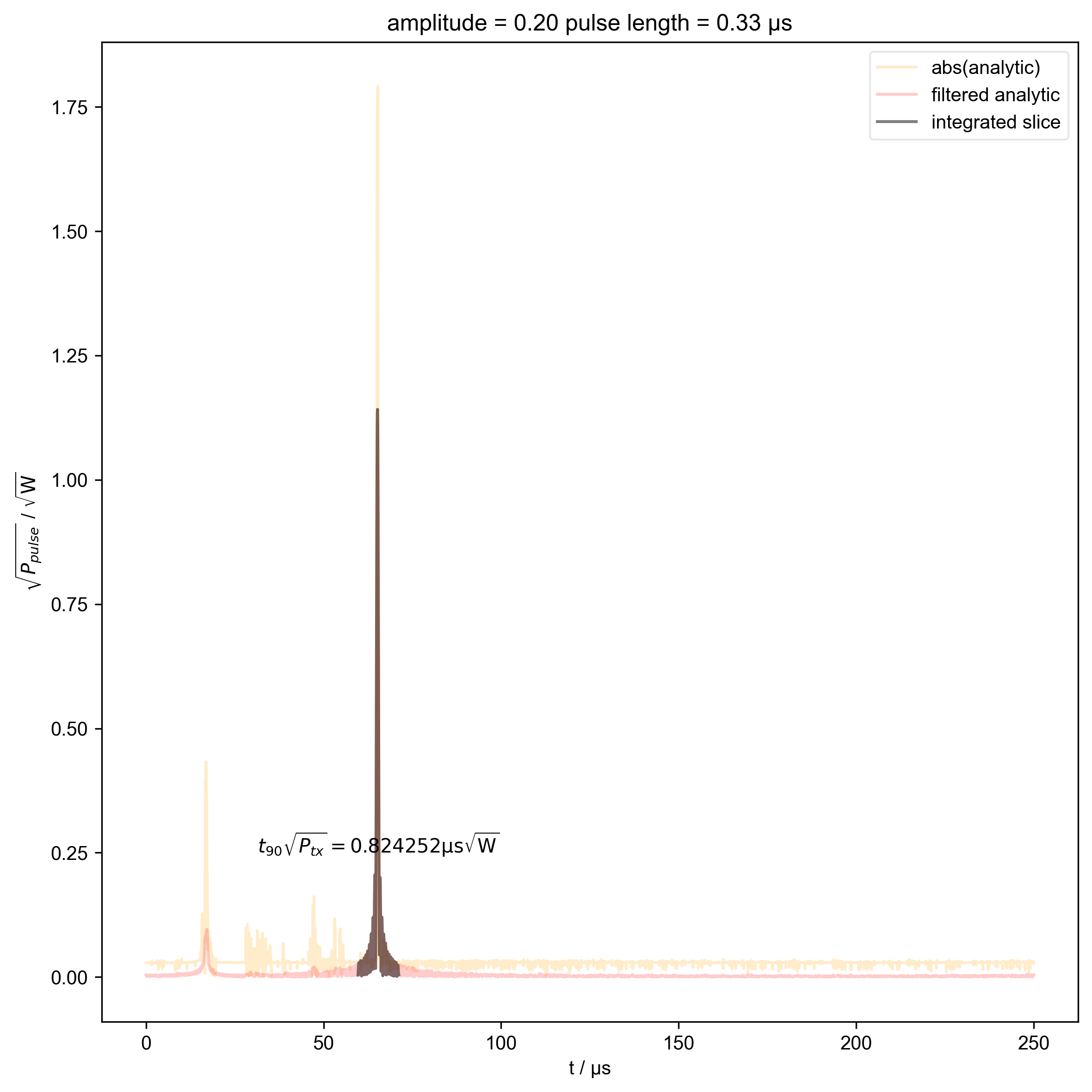
13: amplitude = 0.20 pulse length = 0.33 μs |||μs
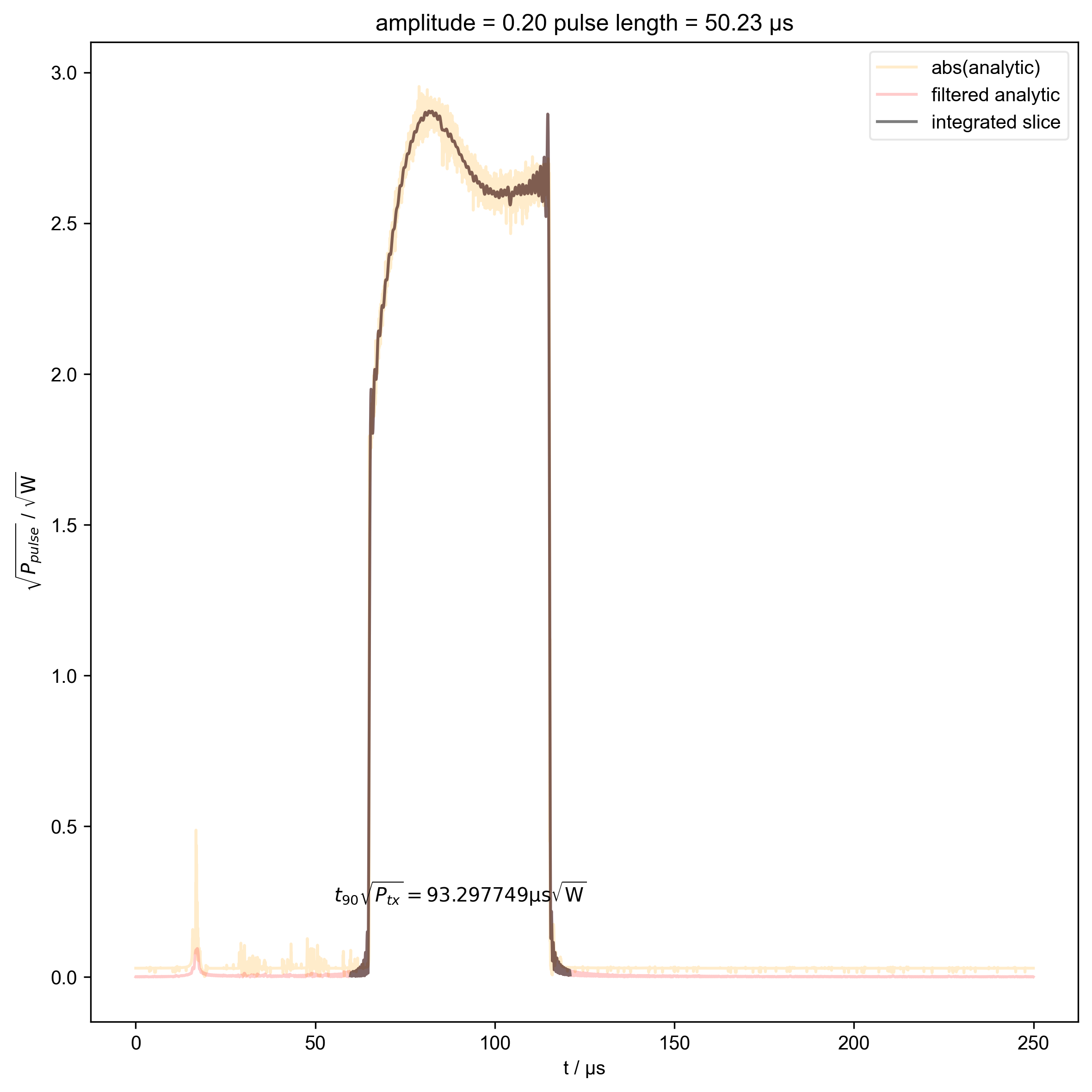
14: amplitude = 0.20 pulse length = 50.23 μs |||μs
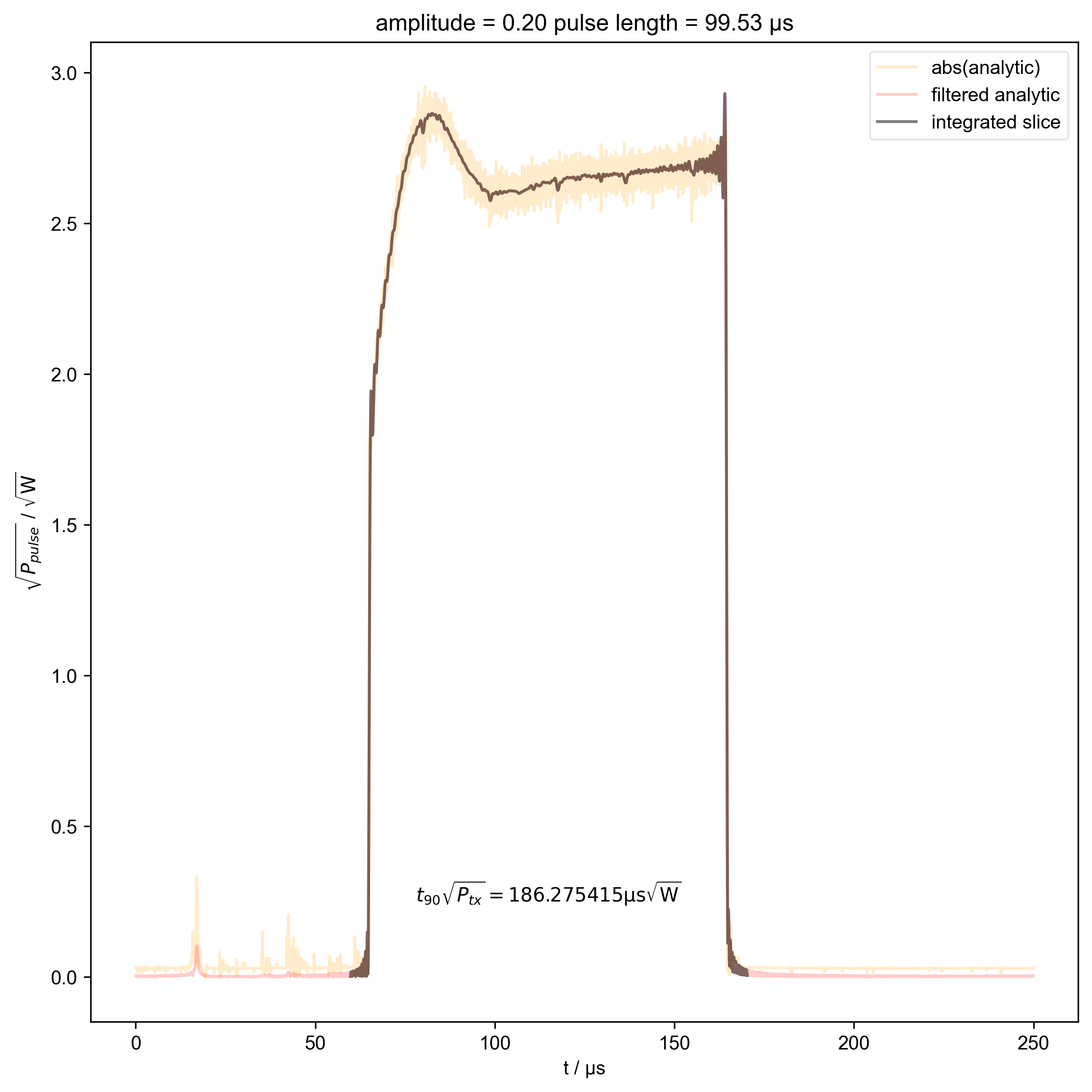
15: amplitude = 0.20 pulse length = 99.53 μs |||μs
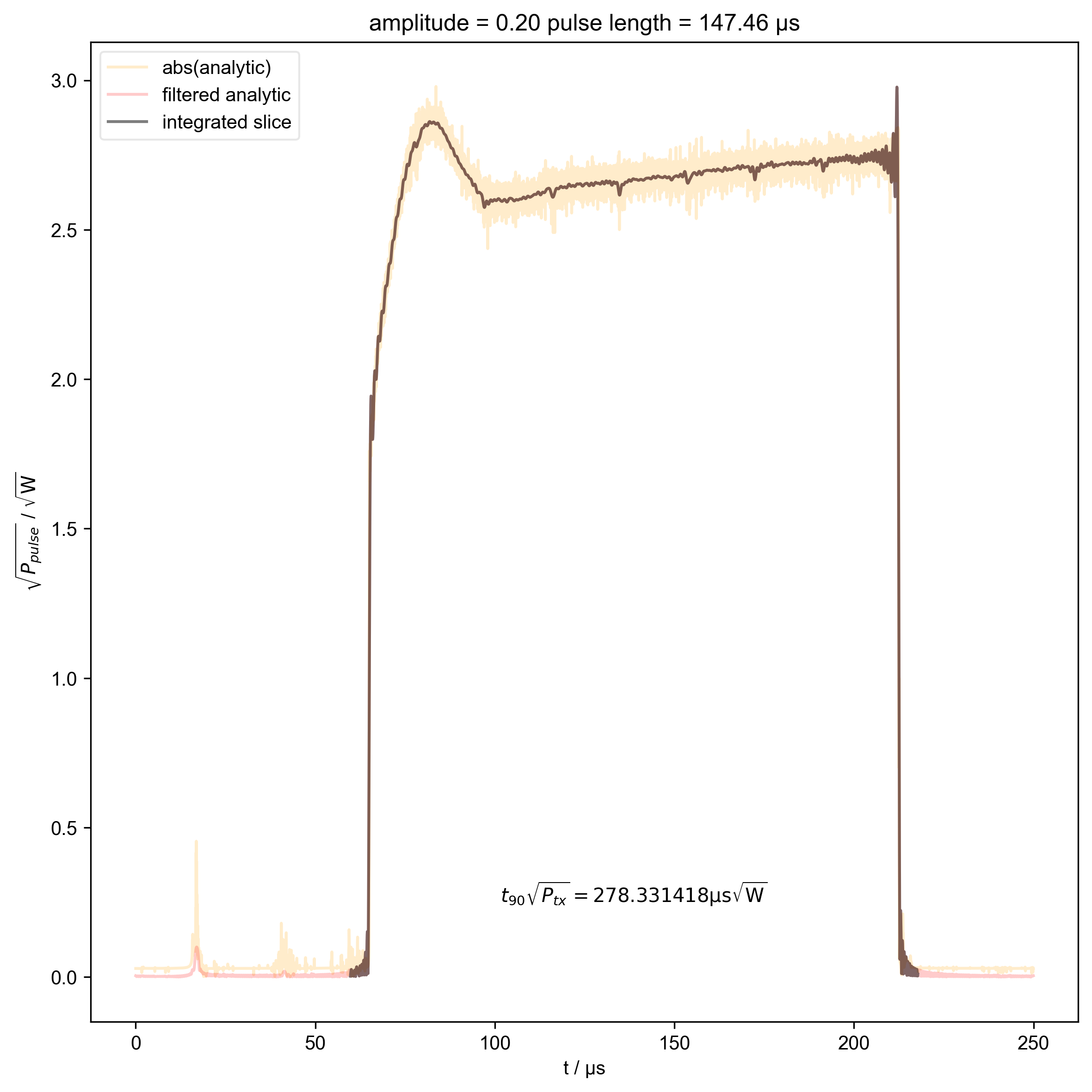
16: amplitude = 0.20 pulse length = 147.46 μs |||μs
17: amplitude = 0.20 Frequency Domain |||('MHz', None)
import pyspecdata as psd
from pyspecProcScripts import find_apparent_anal_freq
import matplotlib.pyplot as plt
import numpy as np
from itertools import cycle
psd.init_logging()
colorcyc_list = plt.rcParams["axes.prop_cycle"].by_key()["color"]
color_cycle = cycle(
colorcyc_list
) # this can be done more than once to spin up multiple lists
V_atten_ratio = 102.2 # attenutation ratio
skip_plots = 33 # diagnostic -- set this to None, and there will be no
# plots
HH_width = 2e6
with psd.figlist_var() as fl:
for filename, nodename in [
(
"240819_test_amp0p05_calib_pulse_calib.h5",
"pulse_calib_5",
),
(
"240819_amp0p1_calib_pulse_calib.h5",
"pulse_calib_2",
),
(
"240819_amp0p2_calib_repeat_pulse_calib.h5",
"pulse_calib_9",
),
]:
d = psd.find_file(
filename, expno=nodename, exp_type="ODNP_NMR_comp/test_equipment"
)
assert (
d.get_prop("postproc_type") == "GDS_capture_v1"
), "The wrong postproc_type was set so you most likely used the wrong script for acquisition"
amplitude = d.get_prop("acq_params")["amplitude"]
fl.basename = f"amplitude = {amplitude:.2f}"
d *= V_atten_ratio # V at output of amplifier
d /= np.sqrt(50) # V/sqrt(R) = sqrt(P_amp)
# {{{ functions that streamline plotting the desired pulse length
# datasets
def switch_to_plot(d, j):
thislen = d.get_prop("programmed_t_pulse")[j] / 1e-6
fl.next(f"pulse length = {thislen:.2f} μs")
def indiv_plots(d, thislabel, thiscolor):
if skip_plots is None:
return
for j in range(len(d["beta"])):
if j % skip_plots == 0:
switch_to_plot(d, j)
fl.plot(
d["beta", j],
alpha=0.2,
color=thiscolor,
label=thislabel,
)
plt.ylabel(r"$\sqrt{P}$ / $\sqrt{\mathrm{W}}$")
# }}}
# {{{ data is already analytic, and downsampled to below 24 MHz
indiv_plots(abs(d), "abs(analytic)", "orange")
d, nu_a, _ = find_apparent_anal_freq(d) # find frequency of signal
d.ft("t")
# {{{ Diagnostic to ensure the frequency properly identified
fl.next("Frequency Domain")
fl.plot(d)
plt.text(
x=0.5,
y=0.5,
s=rf"$\nu_a={nu_a/1e6:0.2f}$ MHz",
transform=plt.gca().transAxes,
)
assert (0 > nu_a * 0.5 * HH_width) or (
0 < nu_a - 0.5 * HH_width
), "unfortunately the region I want to filter includes DC -- this is probably not good, and you should pick a different timescale for your scope so this doesn't happen"
# }}}
# {{{ apply HH frequency filter
d["t" : (None, nu_a - 0.5 * HH_width)] *= 0
d["t" : (nu_a + 0.5 * HH_width, None)] *= 0
# }}}
d.ift("t")
indiv_plots(abs(d), "filtered analytic", "red")
# }}}
# {{{ set up shape of data to drop the calculated beta values in
verify_beta = d.shape.pop("t").alloc(dtype=np.float64)
verify_beta.copy_axes(d)
verify_beta.set_units(r"s√W").set_units("beta", r"s√W")
# }}}
thiscolor = next(color_cycle)
for j in range(len(d["beta"])):
s = d["beta", j]
int_range = abs(s).contiguous(lambda x: x > 0.03 * s.max())[0]
# slightly expand int range to include rising edges
int_range[0] -= 5e-6
int_range[-1] += 5e-6
# {{{ plot the integration range of all pulses prior to integrating.
# Serves as diagnostic to ensure the beta is consistently increasing.
fl.push_marker()
fl.basename = None
fl.next("collect filtered analytic")
fl.plot(abs(s["t":int_range]), alpha=0.3)
fl.pop_marker()
# }}}
verify_beta["beta", j] = abs(s["t":int_range]).integrate(
"t"
).data.item() / np.sqrt(
2
) # tp * sqrt(P_rms)
# {{{ Can't use indiv_plots because we've already indexed the beta
# out and we also want to plot the calculated beta on top
if skip_plots is not None and j % skip_plots == 0:
switch_to_plot(d, j)
fl.plot(
abs(s["t":int_range]),
color="black",
label="integrated slice",
)
plt.ylabel(r"$\sqrt{P_{pulse}}$ / $\sqrt{\mathrm{W}}$")
plt.text(
np.mean(int_range) / 1e-6,
0.25,
r"$t_{90} \sqrt{P_{tx}} = %f \mathrm{μs} \sqrt{\mathrm{W}}$"
% (verify_beta["beta", j].item() / 1e-6),
ha="center",
)
# }}}
# {{{ show what we observe -- how does β vary with the desired β
fl.basename = None # we want to plot all amplitudes together now
fl.next(r"Measured $\beta$ vs programmed $\beta$")
fl.plot(
(verify_beta / 1e-6).set_units("μs√W"),
color=thiscolor,
label="Amplitude = %0.2f" % amplitude,
)
plt.xlabel(r"Programmed $\beta$ / $\mathrm{μs}\sqrt{\mathrm{W}}$")
plt.ylabel(r"Measured $\beta$ / $\mathrm{μs}\sqrt{\mathrm{W}}$")
psd.gridandtick(plt.gca())
# }}}
Total running time of the script: (0 minutes 51.393 seconds)