ProcScripts Example Gallery¶
Below is a gallery of examples using our various functions.
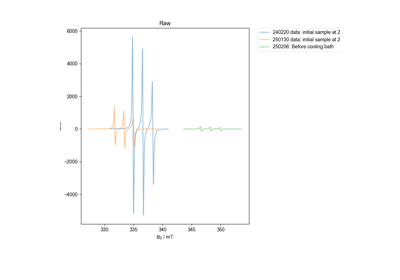
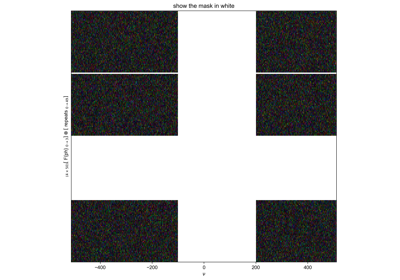
Edit the concentration parameter of a saved HDF5 file
Edit the concentration parameter of a saved HDF5 file
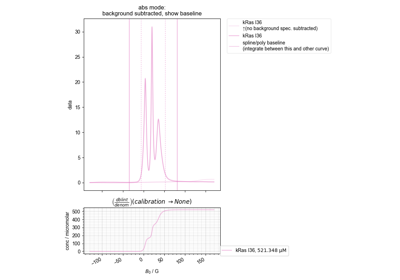
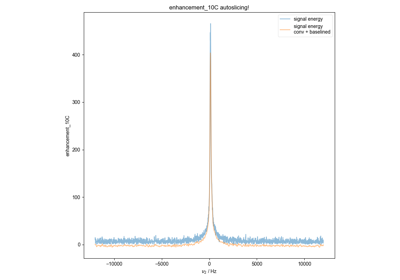
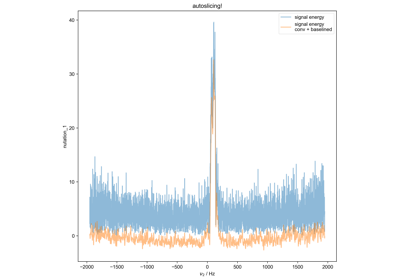
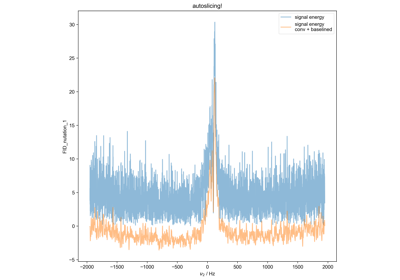
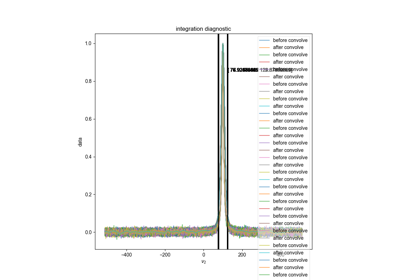
Quantify the Double Integral of an ESR spectra (QESR)
Quantify the Double Integral of an ESR spectra (QESR)
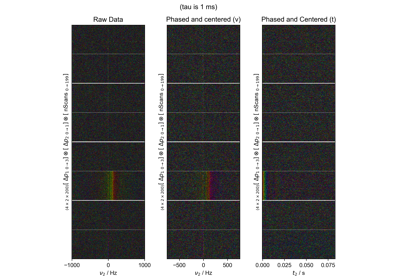
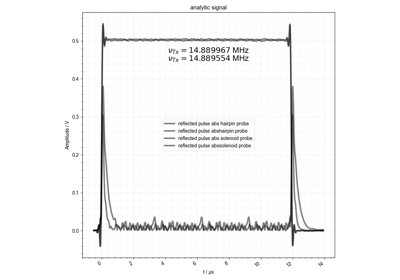
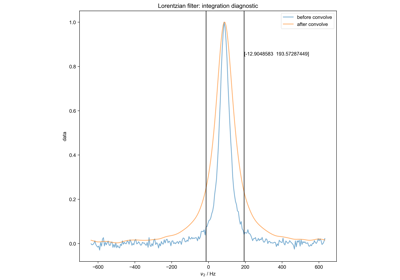
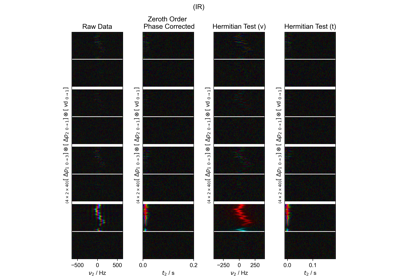
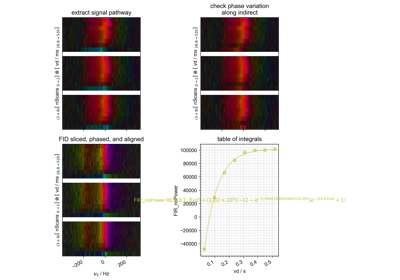
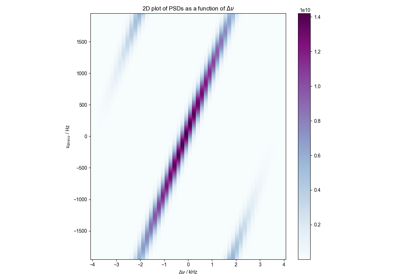
FID from Echo after Phasing and Timing Correction – Challenging Actual Data
FID from Echo after Phasing and Timing Correction -- Challenging Actual Data

sphx_glr_auto_examples_proc_gain.py
"
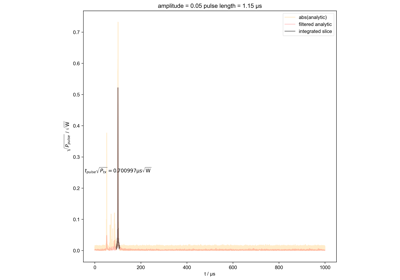
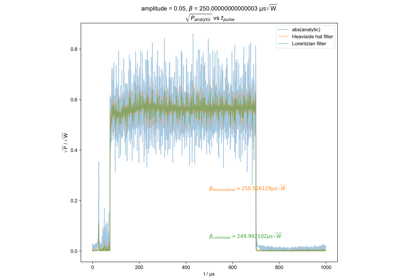
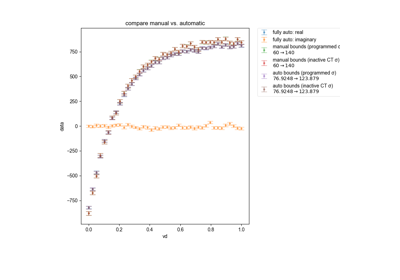
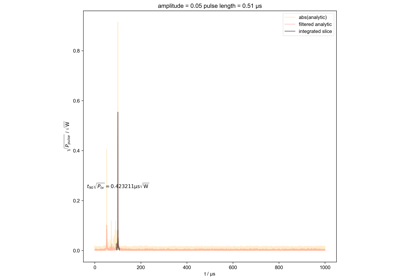
Calculate actual beta as a function of pulse length
Calculate actual beta as a function of pulse length